| A Cross-Species Gene Expression Marker-Based Genetic Map and QTL Analysis in Bambara Groundnut |
|
Bambara groundnut (Vigna subterranea (L.) Verdc.) is an underutilised legume crop, which has long been recognised as a protein-rich and drought-tolerant crop, used extensively in Sub-Saharan Africa. The aim of the study was to identify quantitative trait loci (QTL) involved in agronomic and drought-related traits using an expression marker-based genetic map based on major crop resources developed in soybean. |
|
Chai HH, Ho WK, Graham N, May S, Massawe F, Mayes S.
Genes (Basel). 2017 Feb 22;8(2). pii: E84. doi: 10.3390/genes8020084. AbstractBambara groundnut (Vigna subterranea (L.) Verdc.) is an underutilised legume crop, which has long been recognised as a protein-rich and drought-tolerant crop, used extensively in Sub-Saharan Africa. The aim of the study was to identify quantitative trait loci (QTL) involved in agronomic and drought-related traits using an expression marker-based genetic map based on major crop resources developed in soybean.
The gene expression markers (GEMs) were generated at the (unmasked) probe-pair level after cross-hybridisation of bambara groundnut leaf RNA to the Affymetrix Soybean Genome GeneChip. A total of 753 markers grouped at an LOD (Logarithm of odds) of three, with 527 markers mapped into linkage groups.
From this initial map, a spaced expression marker-based genetic map consisting of 13 linkage groups containing 218 GEMs, spanning 982.7 cM (centimorgan) of the bambara groundnut genome, was developed. Of the QTL detected, 46% were detected in both control and drought treatment populations, suggesting that they are the result of intrinsic trait differences between the parental lines used to construct the cross, with 31% detected in only one of the conditions. The present GEM map in bambara groundnut provides one technically feasible route for the translation of information and resources from major and model plant species to underutilised and resource-poor crops.
See https://www.ncbi.nlm.nih.gov/pubmed/28241413
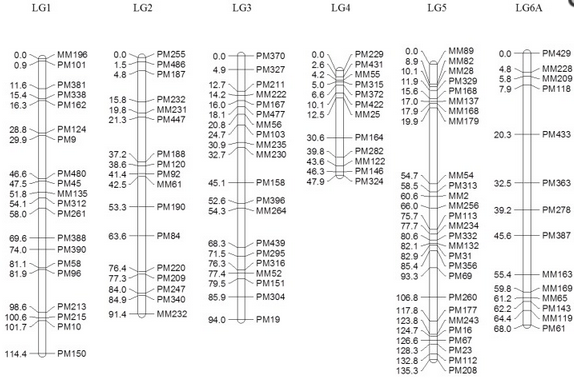
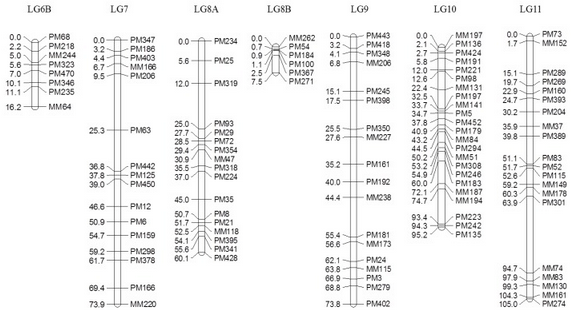
Figure 2: Genetic linkage map of the F5 segregating population in bambara groundnut constructed using gene expression markers (GEMs). Right: positions of markers (cM); left: name of the markers. LG6 and LG8 were divided into subgroups ‘A’ and ‘B’, respectively, based on the association observed in the MLM due to insufficient linkage to complete the map using regression mapping (RM).
|
|
|
|
[ Tin tức liên quan ]___________________________________________________
|


 Curently online :
Curently online :
 Total visitors :
Total visitors :