| Uncovering hidden variation in polyploid wheat |
|
Pasta and bread wheat are polyploid species that carry multiple copies of each gene. Therefore, loss-of-function mutations in one gene copy are frequently masked by functional copies on other genomes. We sequenced the protein coding regions of 2,735 mutant lines and developed a public database including more than 10 million mutations. Researchers and breeders can search this database online, identify mutations in the different copies of their target gene, and request seeds to study gene function or improve wheat varieties |
|
Ksenia V. Krasileva, Hans A. Vasquez-Gross, Tyson Howell, Paul Bailey, Francine Paraiso, Leah Clissold, James Simmonds, Ricardo H. Ramirez-Gonzalez, Xiaodong Wang, Philippa Borrill, Christine Fosker, Sarah Ayling, Andrew L. Phillips, Cristobal Uauy, and Jorge Dubcovsky SignificancePasta and bread wheat are polyploid species that carry multiple copies of each gene. Therefore, loss-of-function mutations in one gene copy are frequently masked by functional copies on other genomes. We sequenced the protein coding regions of 2,735 mutant lines and developed a public database including more than 10 million mutations. Researchers and breeders can search this database online, identify mutations in the different copies of their target gene, and request seeds to study gene function or improve wheat varieties. Mutations are being used to improve the nutritional value of wheat, increase the size of the wheat grains, and generate additional variability in flowering genes to improve wheat adaptation to new and changing environments. AbstractComprehensive reverse genetic resources, which have been key to understanding gene function in diploid model organisms, are missing in many polyploid crops. Young polyploid species such as wheat, which was domesticated less than 10,000 y ago, have high levels of sequence identity among subgenomes that mask the effects of recessive alleles. Such redundancy reduces the probability of selection of favorable mutations during natural or human selection, but also allows wheat to tolerate high densities of induced mutations. Here we exploited this property to sequence and catalog more than 10 million mutations in the protein-coding regions of 2,735 mutant lines of tetraploid and hexaploid wheat. We detected, on average, 2,705 and 5,351 mutations per tetraploid and hexaploid line, respectively, which resulted in 35–40 mutations per kb in each population. With these mutation densities, we identified an average of 23–24 missense and truncation alleles per gene, with at least one truncation or deleterious missense mutation in more than 90% of the captured wheat genes per population. This public collection of mutant seed stocks and sequence data enables rapid identification of mutations in the different copies of the wheat genes, which can be combined to uncover previously hidden variation. Polyploidy is a central phenomenon in plant evolution, and many crop species have undergone recent genome duplication events. Therefore, the general strategy and methods developed herein can benefit other polyploid crops.
See: http://www.pnas.org/content/114/6/E913/F1.expansion.html PNAS February 7 2017; vol.114; no. 6: E913–E921
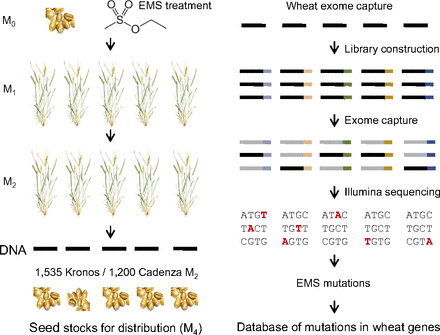
Overview of the development of the sequenced mutant populations. M0 seeds were mutagenized with EMS (resulting in M1 plants), and a single M2 plant was grown from each M1 plant. Genomic DNAs were extracted from the M2 plants, and M3 seeds were obtained from the same plant. M3 seeds were planted in the field to produce M4 seed for distribution. Barcoded sequencing libraries were constructed, used for exome capture, and sequenced by using Illumina. Mutations were identified by using the MAPS pipeline and were deposited in public databases that can be searched online. |
|
|
|
[ Tin tức liên quan ]___________________________________________________
|


 Curently online :
Curently online :
 Total visitors :
Total visitors :