|
Jill M. Bushakra, Douglas W. Bryant, Michael Dossett, Kelly J. Vining, Robert VanBuren, Barbara S. Gilmore, Jungmin Lee, Todd C. Mockler, Chad E. Finn, Nahla V. Bassil
Theoretical and Applied Genetics, August 2015, Volume 128, Issue 8, pp 1631-1646,
http://link.springer.com/article/10.1007/s00122-015-2541-x
Abstract
Key message
We have constructed a densely populated, saturated genetic linkage map of black raspberry and successfully placed a locus for aphid resistance.
Abstract
Black raspberry (Rubus occidentalis L.) is a high-value crop in the Pacific Northwest of North America with an international marketplace. Few genetic resources are readily available and little improvement has been achieved through breeding efforts to address production challenges involved in growing this crop. Contributing to its lack of improvement is low genetic diversity in elite cultivars and an untapped reservoir of genetic diversity from wild germplasm. In the Pacific Northwest, where most production is centered, the current standard commercial cultivar is highly susceptible to the aphid Amphorophora agathonica Hottes, which is a vector for the Raspberry mosaic virus complex. Infection with the virus complex leads to a rapid decline in plant health resulting in field replacement after only 3–4 growing seasons. Sources of aphid resistance have been identified in wild germplasm and are used to develop mapping populations to study the inheritance of these valuable traits. We have constructed a genetic linkage map using single-nucleotide polymorphism and transferable (primarily simple sequence repeat) markers for F1 population ORUS 4305 consisting of 115 progeny that segregate for aphid resistance. Our linkage map of seven linkage groups representing the seven haploid chromosomes of black raspberry consists of 274 markers on the maternal map and 292 markers on the paternal map including a morphological locus for aphid resistance. This is the first linkage map of black raspberry and will aid in developing markers for marker-assisted breeding, comparative mapping with other Rubus species, and enhancing the black raspberry genome assembly.
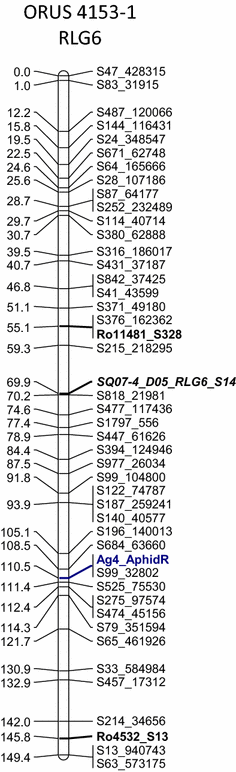
Fig. 2: Rubus linkage group (RLG) 6 for black raspberry mapping population parent ORUS 4153-1. The morphological locus for Ag 4 aphid resistance against the North American large raspberry aphid is shown in blue bold font. The linkage map is constructed of single-nucleotide polymorphic (SNP) loci generated by genotyping by sequencing (GBS) (prefaced with S) and simple sequence repeat (SSR) loci from various Rubus sources (prefaced with Ro, Ri, Rh, Ru, Rub, and SQ). Transferable loci are indicated in bold font; anchor loci for comparisons with other Rubus linkage maps are indicated in bold italic font (color figure online).
|
[ Other News ]___________________________________________________
|