|
CRISPR-Cereal: a guide RNA design tool integrating regulome and genomic variation for wheat, maize and rice
Wednesday, 2021/08/18 | 08:19:47
|
|
Chao He, Hao Liu, Dijun Chen, Wen-Zhao Xie, Mengxin Wang, Yuqi Li, Xin Gong, Wenhao Yan, Ling-Ling Chen Plant Biotechnology Journal; First published: 26 July 2021; https://doi.org/10.1111/pbi.13675
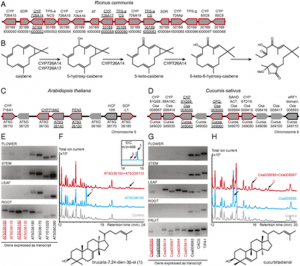
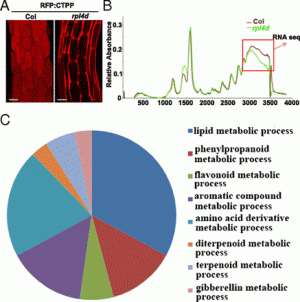
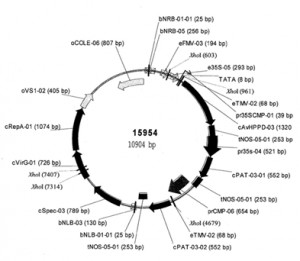
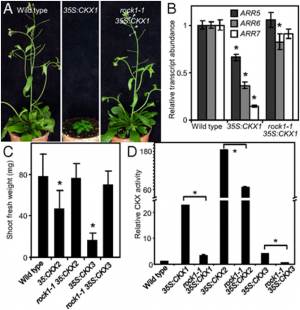
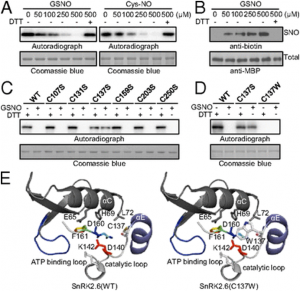
The clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein (Cas) genome editing system (CRISPR-Cas) is revolutionizing agriculture. In this system, a guide sequence that matches to a particular genomic DNA is placed in front of a synthetic RNA that consists of a scaffold sequence necessary for Cas-binding to form a guide RNA (gRNA). gRNA/Cas complex binds to the target DNA that contains a protospacer adjacent motif (PAM) via base-paring and generates a double-strand break (DSB) by Cas protein. Mutations will be created when the DSB cannot be perfectly repaired. Among kinds of Cas variants, Cas9 and Cas12a (also termed Cpf1) are the two major nucleases with highest edit efficiency. NGG (N = A, T, G or C) for SpCas9 from Streptococcus pyogenes, TTTN for Cpf1 from Acidaminococcus or Lachnospiraceae, is necessary for recruiting the nuclease to produce DSBs. The efficiency of CRISPR-Cas is largely determined by the sequence of gRNA and the chromatin status of target region. Guide RNA devotes to direct the CRISPR-Cas to the target editing (on-target) sites and Cas protein binds to open chromatin with higher affinity thus resulting in higher efficiency. In addition, nucleotide polymorphisms in guide sequence greatly affect editing efficiency. To date, a number of CRISPR gRNA design tools have been developed but hardly include other information than base-paring. A gRNA design tool that quickly scans the genome for on-targets and off-targets, and considers chromatin accessibility and single nucleotide polymorphisms (SNPs) at target region is highly demanded, especially for wheat with a gigantic genome. In summary, CRISPR-Cereal integrates regulome information and considers SNPs existed in the candidate gRNAs to promote precise and high-efficient gene editing for wheat, maize and rice.
See: https://onlinelibrary.wiley.com/doi/full/10.1111/pbi.13675
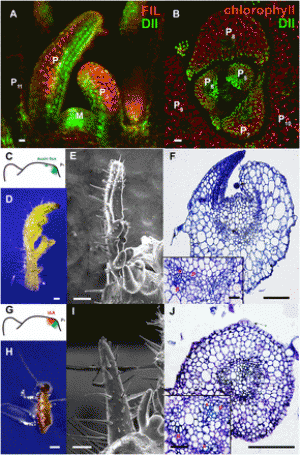
Figure 1: gRNA design using CRISPR-Cereal tool. (a) Regulome and SNP information are available for on-targets identified by CRISPR-Cereal. (b) The crop genomes included in CRISPR-Cereal. (c) The numbers of genome-wide off-targets are increased when allowed mismatches are from zero to five. (d) CRISPR-Cereal basal result page for gRNA designing. (e) The regulome visualization page using GBrowse. (f) The regulome information in 2 kb region around the on-target site. (g) The chromatin accessibility on the upstream 350 bp of OsWOX11. (h) The chromatin accessibility information on TG2 target region. (i) The correlation between mutant frequency and chromatin accessibility in gRNAs target sites. (j) The SNP information on TG7 target region. (k) The workflow of gRNA design using CRISPR-Cereal. |
|
|
|
[ Other News ]___________________________________________________
|


 Curently online :
Curently online :
 Total visitors :
Total visitors :
.jpg)
.jpg)