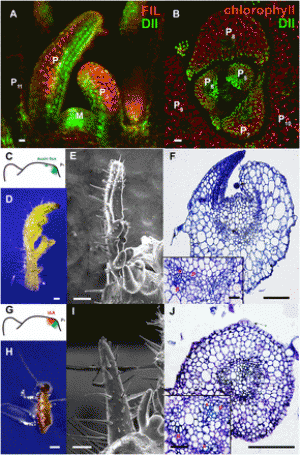
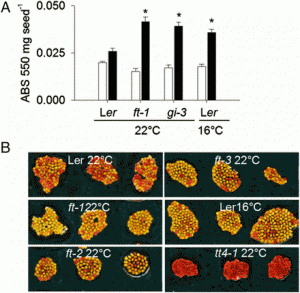
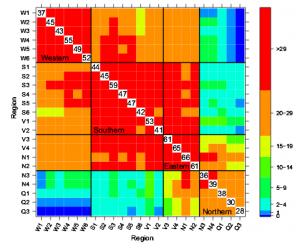
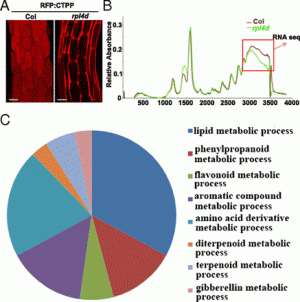
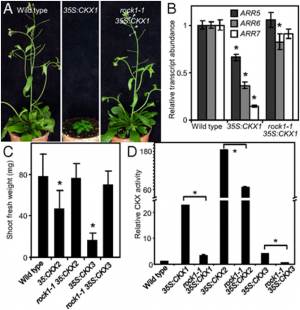
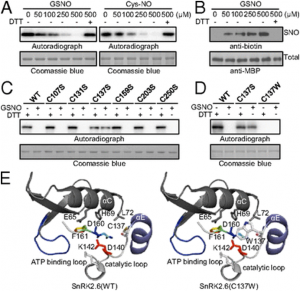
|
Identification and evaluation of quantitative trait loci underlying resistance to multiple HG types of soybean cyst nematode in soybean PI 437655
Tuesday, 2015/01/06 | 08:49:17
|
|
Yongqing Jiao, Tri D. Vuong, Yan Liu, Clinton Meinhardt, Yang Liu, Trupti Joshi, Perry B. Cregan, Dong Xu, J. Grover Shannon, Henry T. Nguyen Theoretical and Applied Genetics, January 2015, Volume 128, Issue 1, pp 15-23, http://link.springer.com/article/10.1007/s00122-014-2409-5 AbstractKey message
We performed QTL analysis for SCN resistance in PI 437655 in two mapping populations, characterized CNV of Rhg1 through whole-genome resequencing and evaluated the effects of QTL pyramiding to enhance resistance. Abstract
Soybean cyst nematode (SCN, Heterodera glycines Ichinohe) is one of the most serious pests of soybean worldwide. PI 437655 has broader resistance to SCN HG types than PI 88788. The objectives of this study were to identify quantitative trait loci (QTL) underlying SCN resistance in PI 437655, and to evaluate the QTL for their contribution to SCN resistance. Two F6:7 recombinant inbred line populations, derived from cv. Williams 82 × PI 437655 and cv. Hutcheson × PI 437655 crosses, were evaluated for resistance to SCN HG types 1.2.5.7 (PA2), 0 (PA3), 1.3.5.6.7 (PA14), and 1.2.3.4.5.6.7 (LY2). The 1,536 SNP array was used to genotype the mapping populations and construct genetic linkage maps. Two significant QTL were consistently mapped on chromosomes (Chr.) 18 and 20 in these two populations. One QTL on Chr. 18, which corresponds to the known Rhg1 locus, contributed resistance to SCN HG types 1.2.5.7, 0, 1.3.5.6.7, and 1.2.3.4.5.6.7 (PA2, PA3, PA14, and LY2, respectively). Copy number variation (CNV) analysis by whole-genome resequencing showed that PI 437655 and PI 88788 had similar CNV at the Rhg1 locus. The QTL on Chr. 20 contributed resistance to SCN HG types 1.3.5.6.7 (PA14) and 1.2.3.4.5.6.7 (LY2). Evaluation of both QTL showed that pyramiding of Rhg1 and the QTL on Chr. 20 significantly improved the resistance to SCN HG types 1.3.5.6.7 (PA14) and 1.2.3.4.5.6.7 (LY2) in both populations. Our studies provided useful information for deploying PI 437655 as a donor for SCN resistance in soybean breeding through marker-assisted selection.
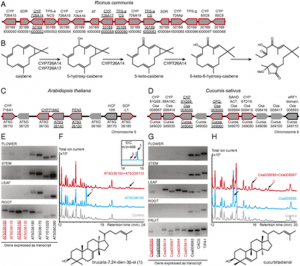
Fig. 2 Copy number variation of the Rhg1 locus detected by CNV-seq software among PI 437655, PI 88788, and cv. Hutcheson by use of whole-genome resequencing. The Yaxis represents log2 ratios; the Xaxis represents genomic positions along chromosome 18 of Wil-liams 82 reference genome. The interval between two black lines indicated the region of Rhg1.The dotrepresents the fold change of copy number variation in comparison with the corresponding soybean line. The red color gradientrepresents Pvalue calculated on each of ratios (color figure online)
|
|
|
|
[ Other News ]___________________________________________________
|


 Curently online :
Curently online :
 Total visitors :
Total visitors :
(22).png)