|
Phytochrome controls alternative splicing to mediate light responses in Arabidopsis
Saturday, 2015/01/03 | 11:18:34
|
|
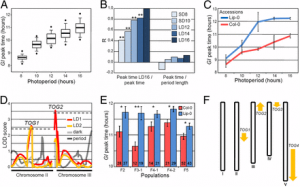
Hiromasa Shikata, Kousuke Hanada, Tomokazu Ushijima, Moeko Nakashima, Yutaka Suzuki, and Tomonao SignificancePlants adapt to their fluctuating environment by monitoring surrounding light conditions through several photoreceptors, such as phytochrome. It is widely believed that upon absorbing red light, phytochrome induces plant light responses by regulating the transcription of numerous target genes. In this study, we provide clear evidence that phytochrome controls not only transcription, but also alternative splicing in Arabidopsis. We reveal that 6.9% of the annotated genes in the Arabidopsis genome undergo rapid changes in their alternative splicing patterns in a red light- and phytochrome-dependent manner. Our results demonstrate that phytochrome simultaneously regulates two different aspects of gene expression, namely transcription and alternative splicing to mediate light responses in plants. AbstractPlants monitor the ambient light conditions using several informational photoreceptors, including red/far-red light absorbing phytochrome. Phytochrome is widely believed to regulate the transcription of light-responsive genes by modulating the activity of several transcription factors. Here we provide evidence that phytochrome significantly changes alternative splicing (AS) profiles at the genomic level in Arabidopsis, to approximately the same degree as it affects steady-state transcript levels. mRNA sequencing analysis revealed that 1,505 and 1,678 genes underwent changes in their AS and steady-state transcript level profiles, respectively, within 1 h of red light exposure in a phytochrome-dependent manner. Furthermore, we show that splicing factor genes were the main early targets of AS control by phytochrome, whereas transcription factor genes were the primary direct targets of phytochrome-mediated transcriptional regulation. We experimentally validated phytochrome-induced changes in the AS of genes that are involved in RNA splicing, phytochrome signaling, the circadian clock, and photosynthesis. Moreover, we show that phytochrome-induced AS changes of SPA1-RELATED 3, the negative regulator of light signaling, physiologically contributed to promoting photomorphogenesis. Finally, photophysiological experiments demonstrated that phytochrome transduces the signal from its photosensory domain to induce light-dependent AS alterations in the nucleus. Taking these data together, we show that phytochrome directly induces AS cascades in parallel with transcriptional cascades to mediate light responses in Arabidopsis.
See: http://www.pnas.org/content/111/52/18781.abstract.html?etoc PNAS December 30, 2014 vol. 111 no. 52: 18781–18786
Fig. 2. A model depicting the early signaling cascades of phytochrome that regulate genome-wide gene expression in response to red light. Red arrows indicate pathways that are regulated by AS; blue arrows indicate pathways subjected to transcriptional regulation. |
|
|
|
[ Other News ]___________________________________________________
|


 Curently online :
Curently online :
 Total visitors :
Total visitors :
.jpg)
.gif)