|
Investigation of terpene diversification across multiple sequenced plant genomes
Friday, 2015/01/09 | 08:51:22
|
|
Alexander M. Boutanaev, Tessa Moses, Jiachen Zi, David R. Nelson, Sam T. Mugford, Reuben J. Peters, and Anne Osbourn Significance
The terpenes are the largest class of plant natural products. This major class of compounds represents tremendous chemical diversity of which only a relatively small fraction has so far been accessed and used by industry. The primary drivers of terpene diversification are terpenoid synthases and cytochromes P450, which synthesize and modify terpene scaffolds. Here, focusing on these two gene families, we investigate terpene synthesis and evolution across 17 sequenced plant genomes. Our analyses shed light on the roots of terpene biosynthesis and diversification in plants. They also reveal that different genomic mechanisms of pathway assembly predominate in eudicots and monocots. Abstract
Plants produce an array of specialized metabolites, including chemicals that are important as medicines, flavors, fragrances, pigments and insecticides. The vast majority of this metabolic diversity is untapped. Here we take a systematic approach toward dissecting genetic components of plant specialized metabolism. Focusing on the terpenes, the largest class of plant natural products, we investigate the basis of terpene diversity through analysis of multiple sequenced plant genomes. The primary drivers of terpene diversification are terpenoid synthase (TS) “signature” enzymes (which generate scaffold diversity), and cytochromes P450 (CYPs), which modify and further diversify these scaffolds, so paving the way for further downstream modifications. Our systematic search of sequenced plant genomes for all TS and CYP genes reveals that distinct TS/CYP gene pairs are found together far more commonly than would be expected by chance, and that certain TS/CYP pairings predominate, providing signals for key events that are likely to have shaped terpene diversity. We recover TS/CYP gene pairs for previously characterized terpene metabolic gene clusters and demonstrate new functional pairing of TSs and CYPs within previously uncharacterized clusters. Unexpectedly, we find evidence for different mechanisms of pathway assembly in eudicots and monocots; in the former, microsyntenic blocks of TS/CYP gene pairs duplicate and provide templates for the evolution of new pathways, whereas in the latter, new pathways arise by mixing and matching of individual TS and CYP genes through dynamic genome rearrangements. This is, to our knowledge, the first documented observation of the unique pattern of TS and CYP assembly in eudicots and monocots.
See: http://www.pnas.org/content/112/1/E81.abstract.html?etoc PNAS January 6, 2015 vol. 112 no. 1: E81–E88
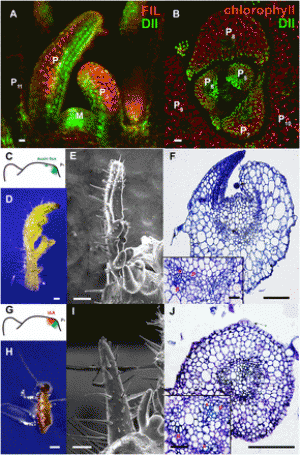
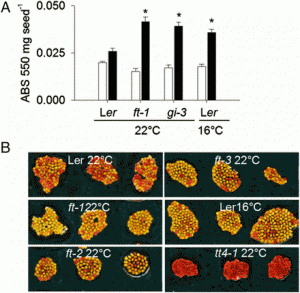
Fig. 2. Functional analysis of R. communis TPS-a/CYP726, A. thaliana TTC/CYP716, and C. sativus TTC/CYP81 gene pairs. (A) Gene components of the R. communis diterpenoid cluster region consisting of 16 genes. AT, acyl-transferase; CS, casbene synthase; NCS, neo-cembrene synthase; SDR, short-chain alcohol dehydrogenase/reductase. (B) Reactions catalyzed by CYP726A14 (or CYP726A17) with casbene and potential role in castor bean diterpenoid biosynthesis. (C and D) Gene components of the A. thaliana PEN3 cluster region on chromosome 5 (C) and C. sativus Csa008595 cluster region on chromosome 6 (D). The TTC/CYP716 and TTC/CYP81 gene pairs identified in our bioinformatics analysis are underlined. Arrows boxed in: red, candidate cluster genes; black, gene adjacent to cluster. (E,G) Qualitative RT-PCR analysis of candidate cluster genes, gene adjacent to the cluster and two housekeeping genes in different tissues of A. thaliana (E) and C. sativus (G). Candidate genes showing tissue-specific coexpression are highlighted in red. Gene pairs identified in our bioinformatics analysis are underlined. (F and H) GC-MS analysis of N. benthamiana leaf extracts transiently expressing AT5G36150 with (red) or without (blue) AT5G36110 (F) or Csa008595 with (red) or without (blue) Csa008597 (H), and an untransformed leaf control (gray). Peaks unique to each chromatogram are pointed with arrows. EIC, extracted ion chromatogram. EI-MS corresponding to the peaks are given in the SI Appendix, Fig. S2 D and I. Structures of the TTC cyclization product are given at the bottom. |
|
|
|
[ Other News ]___________________________________________________
|


 Curently online :
Curently online :
 Total visitors :
Total visitors :
(18).png)