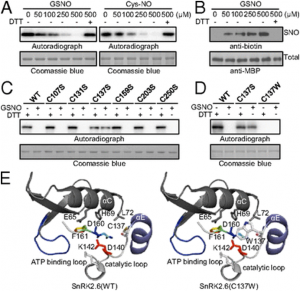
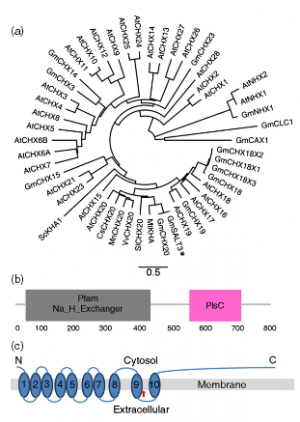
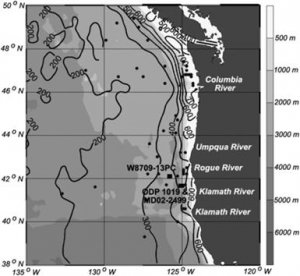
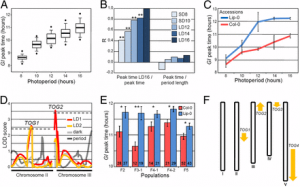
|
Genome diversity of tuber-bearing Solanum uncovers complex evolutionary history and targets of domestication in the cultivated potato
Monday, 2017/12/25 | 08:18:05
|
|
Michael A. Hardigan, F. Parker E. Laimbeer, Linsey Newton, Emily Crisovan, John P. Hamilton, Brieanne Vaillancourt, Krystle Wiegert-Rininger, Joshua C. Wood, David S. Douches, Eva M. Farré, Richard E. Veilleux, and C. Robin Buell. PNAS 2017 vol.114, no.46: E9999–E10008 SignificanceWorldwide, potato is the third most important crop grown for direct human consumption, but breeders have struggled to produce new varieties that outperform those released over a century ago, as evidenced by the most widely grown North American cultivar (Russet Burbank) released in 1876. Despite its importance, potato genetic diversity at the whole-genome level remains largely unexplored. Analysis of cultivated potato and its wild relatives using modern genomics approaches can provide insight into the genomic diversity of extant germplasm, reveal historic introgressions and hybridization events, and identify genes targeted during domestication that control variance for agricultural traits, all critical information to address food security in 21st century agriculture. AbstractCultivated potatoes (Solanum tuberosum L.), domesticated from wild Solanum species native to the Andes of southern Peru, possess a diverse gene pool representing more than 100 tuber-bearing relatives (Solanum section Petota). A diversity panel of wild species, landraces, and cultivars was sequenced to assess genetic variation within tuber-bearing Solanum and the impact of domestication on genome diversity and identify key loci selected for cultivation in North and South America. Sequence diversity of diploid and tetraploid S. tuberosum exceeded any crop resequencing study to date, in part due to expanded wild introgressions following polyploidy that captured alleles outside of their geographic origin. We identified 2,622 genes as under selection, with only 14–16% shared by North American and Andean cultivars, showing that a limited gene set drove early improvement of cultivated potato, while adaptation of upland (S. tuberosum group Andigena) and lowland (S. tuberosum groups Chilotanum and Tuberosum) populations targeted distinct loci. Signatures of selection were uncovered in genes controlling carbohydrate metabolism, glycoalkaloid biosynthesis, the shikimate pathway, the cell cycle, and circadian rhythm. Reduced sexual fertility that accompanied the shift to asexual reproduction in cultivars was reflected by signatures of selection in genes regulating pollen development/gametogenesis. Exploration of haplotype diversity at potato’s maturity locus (StCDF1) revealed introgression of truncated alleles from wild species, particularly S. microdontum in long-day–adapted cultivars. This study uncovers a historic role of wild Solanum species in the diversification of long-day–adapted tetraploid potatoes, showing that extant natural populations represent an essential source of untapped adaptive potential.
See: http://www.pnas.org/content/114/46/E9999.full
Figure 3: Wild Solanum species introgressions in cultivated potato. (A) Fraction of assessed genome sequences (5-kb windows) with introgressions from individual wild species in diploid landraces, tetraploid landraces, and cultivars. (B) Map of wild species introgressions on potato chromosome 11 for diploid landraces, tetraploid landraces, and cultivars. Color codes for species introgressions are blue, ambiguous/multiple taxa; red, S. microdontum; green, S. candolleanum; orange, Solanum sparsipilum; purple, S. leptophyes; pink, S. raphanifolium; gold, S. brevicaule; brown, S. medians; navy, S. chacoense; dark red, S. berthaultii; and light green, Solanum infundibuliforme. All 12 chromosomes, including names of accessions |
|
|
|
[ Other News ]___________________________________________________
|


 Curently online :
Curently online :
 Total visitors :
Total visitors :
(46).png)