|
A second VrPGIP1 allele is associated with bruchid resistance (Callosobruchus spp.) in wild mungbean (Vigna radiata var. sublobata) accession ACC41.
Wednesday, 2019/11/13 | 08:23:23
|
|
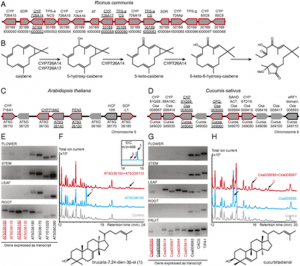
Kaewwongwal A, Liu C3, Somta P, Chen J, Tian J, Yuan X, Chen X. Mol Genet Genomics. 2019 Nov 8. doi: 10.1007/s00438-019-01619-y. [Epub ahead of print] AbstractTwo bruchid species, azuki bean weevil (Callosobruchus chinensis L.) and cowpea weevil (Callosobruchus maculatus F.), are the most important insect pests of mungbean [Vigna radiata (L.) Wilczek] after harvest. Improving bruchid resistance is a major goal for mungbean breeders. Bruchid resistance in mungbean is controlled by a single major locus, Br. The tightly linked VrPGIP1 and VrPGIP2, which encode polygalacturonase-inhibiting proteins (PGIPs), are the candidate genes at the Br locus associated with bruchid resistance. One VrPGIP1 resistance allele and two VrPGIP2 resistance alleles have been identified. In this study, we fine-mapped the bruchid-resistance genes in wild mungbean (V. radiata var. sublobata) accession ACC41 using the F2 population (574 individuals) derived from the 'Kamphaeng Saen 2' (susceptible) × ACC41 (resistant) cross. A QTL analysis indicated that the resistance to the azuki bean weevil and cowpea weevil in ACC41 is controlled by a major QTL (qBr5.1) and a minor QTL (qBr5.2), which are only 0.3 cM apart. qBr5.1 and qBr5.2 accounted for about 82% and 2% of the resistance variation in the F2 population, respectively. qBr5.1 was mapped to a 237.35-kb region on mungbean chromosome 5 containing eight annotated genes, including VrPGIP1 and VrPGIP2. An examination of the ACC41 VrPGIP1 and VrPGIP2 sequences revealed a new allele for VrPGIP1 (i.e., VrPGIP1-2). Compared with the wild-type sequence, VrPGIP1-2 has five SNPs, of which four cause amino acid changes (residues 125, 129, 188, and 336). A protein sequence analysis indicated that residues 125 and 129 in VrPGIP1-2 are in a β-sheet B1 region, whereas residues 188 and 336 are in a C10-helix region and at the end of the C-terminal region, respectively. Because the β-sheet B1 region is important for interactions with polygalacturonase (PG), residues 125 and 129 in VrPGIP1-2 likely contribute to bruchid resistance by inhibiting PG. Our results imply that VrPGIP1-2 is associated with the bruchid resistance of wild mungbean accession ACC41. This new resistance allele may be useful for breeding mungbean varieties exhibiting durable bruchid resistance.
|
|
|
|
[ Other News ]___________________________________________________
|


 Curently online :
Curently online :
 Total visitors :
Total visitors :