|
Arabidopsis SAURs are critical for differential light regulation of the development of various organs
Sunday, 2016/05/29 | 06:34:25
|
|
Ning Sun, Jiajun Wang, Zhaoxu Gao, Jie Dong, Hang He, William Terzaghi, Ning Wei, Xing Wang Deng, and Haodong Chen SignificanceVarious plant organs respond differentially to environmental signals so that plants can adapt to dynamic environments without movement. Light is a key environmental factor mediating multiple plant developmental processes. For example, it induces cotyledon expansion but inhibits hypocotyl elongation when plants emerge from soil. Although this opposite regulation is crucial for plant survival and has been described for decades, the underlying mechanism is still elusive. In this study, we demonstrated that temporal–spatial expression of a group of Small Auxin Up RNAs (SAURs) is regulated by light through auxin and phytochrome-interacting factors, and these SAURs further mediate the differential growth of cotyledons and hypocotyls. Thus, this study provides a molecular mechanism explaining how light differentially regulates the growth of various plant organs. AbstractDuring deetiolation of Arabidopsis seedlings, light promotes the expansion of cotyledons but inhibits the elongation of hypocotyls. The mechanism of this differential regulation of cell enlargement is unclear. Our organ-specific transcriptomic analysis identified 32 Small Auxin Up RNA (SAUR) genes whose transcripts were light-induced in cotyledons and/or repressed in hypocotyls. We therefore named these SAURs as lirSAURs. Both overexpression and mutation analyses demonstrated that lirSAURs could promote cotyledon expansion and opening and enhance hypocotyl elongation, possibly by inhibiting phosphatase activity of D-clade type 2C protein phosphatases (PP2C-Ds). Light reduced auxin levels to down-regulate the expression of lirSAURs in hypocotyls. Further, phytochrome-interacting factors (PIFs) were shown to directly bind the genes encoding these SAURs and differentially regulate their expression in cotyledons and hypocotyls. Together, our study demonstrates that light mediates auxin levels and PIF stability to differentially regulate the expression of lirSAURs in cotyledons and hypocotyls, and these lirSAURs further mediate the differential growth of these two organs.
See: http://www.pnas.org/content/113/21/6071.full PNAS May 24 2016; vol.111; no.21: 6071–6076
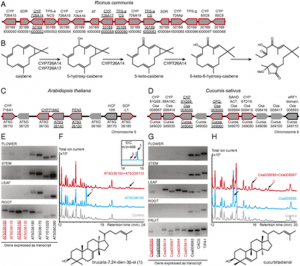
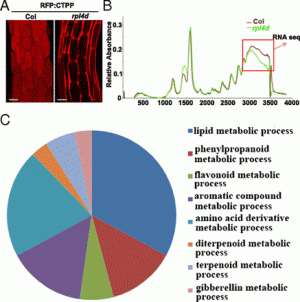
Figure 1: Many SAUR family genes are up-regulated in cotyledons and/or down-regulated in hypocotyls during the dark-to-light transition. (A) Morphological comparison of 4-d-old WT Arabidopsis seedlings grown in continuous white light (L), continuous darkness (D), or transferred from darkness to light and then kept in white light for 1 h (DL 1h), 6 h (DL 6h), and 24 h (DL 24h). (Scale bar: 2 mm.) (B) Graphic diagram of RNA-Seq samples. Four-day-old dark-grown seedlings were exposed to white light for 0 h, 1 h, and 6 h, and cotyledons and hypocotyls were dissected at each time point. The Bottom diagram indicates the dissection sites of the seedlings. (C) Venn diagram showing numbers of light-responsive genes at 1 h (DL1h/D) or 6 h (DL6h/D) in cotyledons (C), or hypocotyls (H), respectively. Subgroup numbers are labeled in circles. (D) Heat map of the genes in subgroup 2, subgroup 5, and subgroup 8 of C. The red box marks the genes oppositely regulated by light in cotyledons and hypocotyls. (E) Expression patterns of representatives of four classes of SAUR genes. (F) Pie chart showing the numbers of genes in each class and their percentages in the SAUR gene family. |
|
|
|
[ Other News ]___________________________________________________
|


 Curently online :
Curently online :
 Total visitors :
Total visitors :
.gif)