|
Pangenome-Wide Association Study and Transcriptome Analysis Reveal a Novel QTL and Candidate Genes Controlling both Panicle and Leaf Blast Resistance in Rice
Wednesday, 2024/07/31 | 08:33:37
|
|
Jian Wang, Haifei Hu, Xianya Jiang, Shaohong Zhang, Wu Yang, Jingfang Dong, Tifeng Yang, Yamei Ma, Lian Zhou, Jiansong Chen, Shuai Nie, Chuanguang Liu, Yuese Ning, Xiaoyuan Zhu, Bin Liu, Jianyuan Yang, Junliang Zhao Rice (N Y); 2024 Apr 12; 17(1):27. doi: 10.1186/s12284-024-00707-x. AbstractCultivating rice varieties with robust blast resistance is the most effective and economical way to manage the rice blast disease. However, rice blast disease comprises leaf and panicle blast, which are different in terms of resistance mechanisms. While many blast resistant rice cultivars were bred using genes conferring resistance to only leaf or panicle blast, mining durable and effective quantitative trait loci (QTLs) for both panicle and leaf blast resistance is of paramount importance. In this study, we conducted a pangenome-wide association study (panGWAS) on 9 blast resistance related phenotypes using 414 international diverse rice accessions from an international rice panel. This approach led to the identification of 74 QTLs associated with rice blast resistance. One notable locus, qPBR1, validated in a F4:5 population and fine-mapped in a Heterogeneous Inbred Family (HIF), exhibited broad-spectrum, major and durable blast resistance throughout the growth period. Furthermore, we performed transcriptomic analysis of 3 resistant and 3 sensitive accessions at different time points after infection, revealing 3,311 differentially expressed genes (DEGs) potentially involved in blast resistance. Integration of the above results identified 6 candidate genes within the qPBR1 locus, with no significant negative effect on yield. The results of this study provide valuable germplasm resources, QTLs, blast response genes and candidate functional genes for developing rice varieties with enduring and broad-spectrum blast resistance. The qPBR1, in particular, holds significant potential for breeding new rice varieties with comprehensive and durable resistance throughout their growth period.
See https://pubmed.ncbi.nlm.nih.gov/38607544/
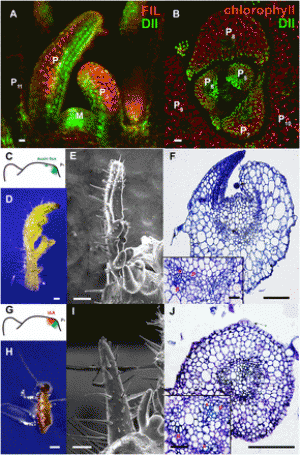
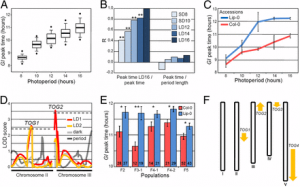
Identification of QTLs for panicle, leaf and seedling blast resistance. A Manhattan plots of the GWAS with panicle blast resistance (PBR) in field. B Manhattan plots of the GWAS with leaf blast resistance (LBR) in field with all accessions. C–I Manhattan plots of the GWAS with seedling blast resistance (LB_ISO11.121, LB_ISO08T19, LB_ISO11.882, LB_ISO11.1093, LB_ISO13.227, LB_ISO14.42, LB_ISO98.288) to a strain ISO11.121, ISO08T19, ISO11.882, ISO11.1093, ISO13.227, ISO14.42, ISO98.288 at seedling stage in green house. J Manhattan plots of the GWAS with leaf blast resistance in field without extremely resistant plants. K–T QQ plot for the GWAS. y-axis: observed -log10(p) and x-axis: expected -log10(p) under the assumption that p follows a uniform [0,1] distribution. The red lines and gray region show the 95% confidence interval for the QQ plot under the null hypothesis of no association between the SNP and the trait
|
|
|
|
[ Other News ]___________________________________________________
|


 Curently online :
Curently online :
 Total visitors :
Total visitors :
(277).png)